空间插值或预测模型的准确性至关重要,因为它决定了插值值的质量。与空间预测的准确性评估相比,开发空间预测模型要容易得多,但是通常情况下,空间预测模型仍然未知。
空间预测质量的评价措施是: 具有 kriging 方差的残差的平均误差 (ME),平均绝对误差 (MAE),均方根误差 (RMSE) 和 均方偏差比 (MSDR) ,可以计算为:
在本练习中,我们将使用以下三种方法评估普通克里金法预测的质量:
- 交叉验证
- 使用独立数据集进行验证
- 空间不确定性的条件模拟
原文链接:Geospatial Data Science with R
全部机翻,如有错误,以你的感觉为准。
交叉验证
交叉验证是一种重新抽样的过程,用于在有限的数据样本上评估模型。它优于残差评价。两种主要的交叉验证技术通常用于模型评估: 1) K-fold 交叉验证(K-fold cross validation); 2)留一交叉验证(Leave-one-out cross validation)。
在 K 折交叉验证中,将数据集随机分为测试集和训练集 k 个不同的时间,并重复进行模型进化 k 次。每次,将 k 个子集中的一个作为测试集,将其他 k-1 子集放在一起形成训练集。然后计算所有 k 个试验的平均误差。该方法的一种变体是将数据随机分为 k 个不同时间的测试和训练集。
在留一交叉验证中,K 等于 N,集合中的数据点的数量。对除一个点以外的所有数据训练模型,并对该点进行预测。最终,使用所有其他观测值分别在每个观测点进行模型预测,并计算平均误差并将其用于评估模型。
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
| library(plyr)
library(dplyr)
library(gstat)
library(raster)
library(ggplot2)
library(car)
library(classInt)
library(RStoolbox)
library(spatstat)
library(dismo)
library(fields)
library(gridExtra)
# Define data folder
dataFolder<-"D:\\Env\\DATA_08\\"
train<-read.csv(paste0(dataFolder,"train_data.csv"), header= TRUE)
|
数据变换
幂变换使用 Box和Cox (1964) 的最大似然方法来选择针对正态性的单变量或多变量响应的变换。首先,我们必须使用 car 包的 powerTransform() 函数计算适当的转换参数,然后使用此参数使用 bcPower() 函数对数据进行转换。
1
2
3
4
5
6
7
| powerTransform(train$SOC)
# Estimated transformation parameter
# train$SOC
# 0.2523339
train$SOC.bc<-bcPower(train$SOC, 0.2523339)
coordinates(train) = ~x+y
|
变异函数建模
1
2
3
4
5
6
7
8
9
10
| # Variogram
v<-variogram(SOC.bc~ 1, data = train, cloud=F)
# Intial parameter set by eye esitmation
m<-vgm(1.5,"Exp",40000,0.5)
# least square fit
m.f<-fit.variogram(v, m)
m.f
# model psill range
# 1 Nug 0.5165678 0.00
# 2 Exp 1.0816886 82374.23
|
K-fold 交叉验证
我们将使用 k-fold 交叉验证对模型进行评估。我们将使用 krige.cv() 函数。
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
| cv<-krige.cv(SOC.bc ~ 1,
train, # data
model = m.f, # fitted varigram model
nfold=10) # five-fold cross validation
summary(cv)
# Object of class SpatialPointsDataFrame
# Coordinates:
# min max
# x -1246454 83927.82
# y 1019863 2526240.55
# Is projected: NA
# proj4string : [NA]
# Number of points: 368
# Data attributes:
# var1.pred var1.var observed residual
# Min. :0.1761 Min. :0.8063 Min. :-1.499 Min. :-3.188626
# 1st Qu.:1.4505 1st Qu.:0.9752 1st Qu.: 1.149 1st Qu.:-0.721998
# Median :2.0100 Median :1.0540 Median : 1.974 Median :-0.056187
# Mean :1.9901 Mean :1.0600 Mean : 1.981 Mean :-0.008895
# 3rd Qu.:2.5575 3rd Qu.:1.1274 3rd Qu.: 2.919 3rd Qu.: 0.713665
# Max. :3.9450 Max. :1.4779 Max. : 5.423 Max. : 2.936493
# zscore fold
# Min. :-3.286167 Min. : 1.000
# 1st Qu.:-0.687623 1st Qu.: 3.000
# Median :-0.055530 Median : 6.000
# Mean :-0.007057 Mean : 5.639
# 3rd Qu.: 0.693822 3rd Qu.: 8.000
# Max. : 2.994699 Max. :10.000
|
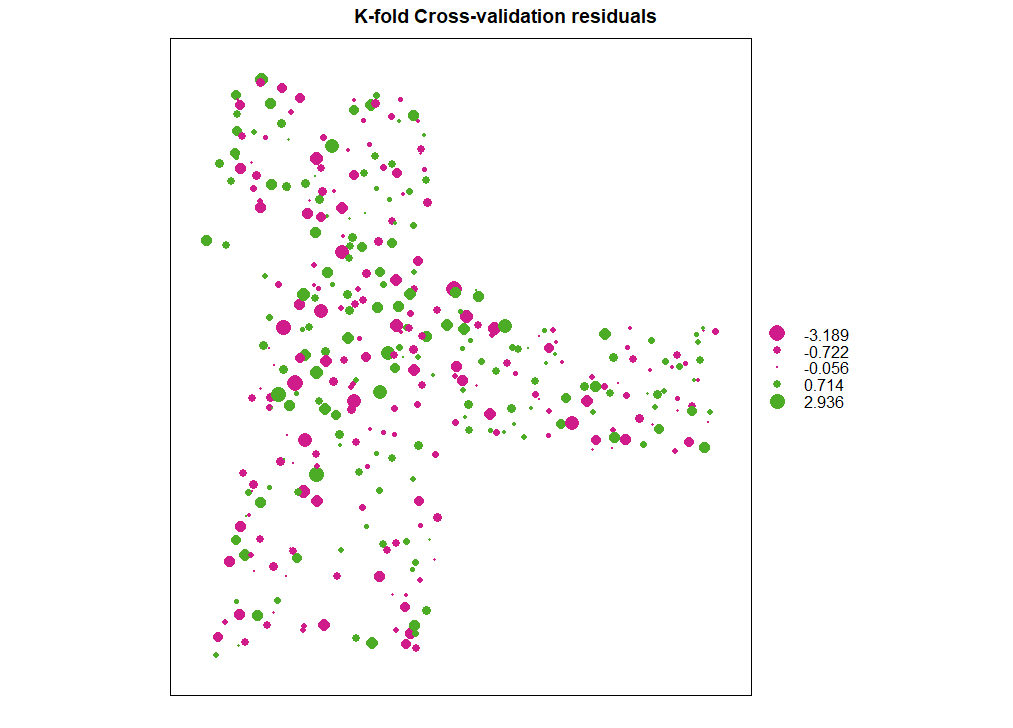
残差图
1
| bubble(cv, zcol = "residual", maxsize = 2.0, main = "K-fold Cross-validation residuals")
|
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
| # Mean Error (ME)
ME<-round(mean(cv$residual),3)
# Mean Absolute Error
MAE<-round(mean(abs(cv$residual)),3)
# Root Mean Squre Error (RMSE)
RMSE<-round(sqrt(mean(cv$residual^2)),3)
# Mean Squared Deviation Ratio (MSDR)
MSDR<-mean(cv$residual^2/cv$var1.var)
ME
# [1] -0.009
MAE
# [1] 0.826
RMSE
# [1] 1.04
MSDR
# [1] 1.03143
|
实际值 vs 预测值: 线性回归
比较实际值与预测值的另一种方法是在它们之间进行线性回归。理想情况下,这将是一条1:1 的线: 截距为 0 (无偏差),斜率设置为 1 (增益相等)。
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
| lm.cv <- lm(cv$var1.pred ~ cv$observed)
summary(lm.cv)
# Call:
# lm(formula = cv$var1.pred ~ cv$observed)
# Residuals:
# Min 1Q Median 3Q Max
# -1.70179 -0.40920 -0.03429 0.44986 2.20694
# Coefficients:
# Estimate Std. Error t value Pr(>|t|)
# (Intercept) 1.2902 0.0602 21.43 <2e-16 ***
# cv$observed 0.3533 0.0255 13.85 <2e-16 ***
# ---
# Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
# Residual standard error: 0.6279 on 366 degrees of freedom
# Multiple R-squared: 0.3439, Adjusted R-squared: 0.3422
# F-statistic: 191.9 on 1 and 366 DF, p-value: < 2.2e-16
|
1:1 绘图
留一交叉验证
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
| cv.LOOCV<-krige.cv(SOC.bc ~ 1,
train, # data
model = m.f) # fitted varigram model
summary(cv.LOOCV)
# Object of class SpatialPointsDataFrame
# Coordinates:
# min max
# x -1246454 83927.82
# y 1019863 2526240.55
# Is projected: NA
# proj4string : [NA]
# Number of points: 368
# Data attributes:
# var1.pred var1.var observed residual
# Min. :0.1107 Min. :0.8041 Min. :-1.499 Min. :-3.188887
# 1st Qu.:1.4400 1st Qu.:0.9591 1st Qu.: 1.149 1st Qu.:-0.708533
# Median :1.9942 Median :1.0281 Median : 1.974 Median :-0.008027
# Mean :1.9824 Mean :1.0382 Mean : 1.981 Mean :-0.001238
# 3rd Qu.:2.5319 3rd Qu.:1.1016 3rd Qu.: 2.919 3rd Qu.: 0.723783
# Max. :3.9177 Max. :1.4022 Max. : 5.423 Max. : 2.934362
# zscore fold
# Min. :-3.286447 Min. : 1.00
# 1st Qu.:-0.698335 1st Qu.: 92.75
# Median :-0.008109 Median :184.50
# Mean :-0.000655 Mean :184.50
# 3rd Qu.: 0.721153 3rd Qu.:276.25
# Max. : 2.992528 Max. :368.00
|
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
| # Mean Error (ME)
ME.LOOCV<-round(mean(cv.LOOCV$residual),3)
# Mean Absolute Error
MAE.LOOCV<-round(mean(abs(cv.LOOCV$residual)),3)
# Root Mean Squre Error (RMSE)
RMSE.LOOCV<-round(sqrt(mean(cv.LOOCV$residual^2)),3)
# Mean Squared Deviation Ratio (MSDR)
MSDR.LOOCV<-mean(cv.LOOCV$residual^2/cv$var1.var)
ME.LOOCV
# [1] -0.001
MAE.LOOCV
# [1] 0.832
RMSE.LOOCV
# [1] 1.042
MSDR.LOOCV
# [1] 1.033859
|
使用独立数据集进行验证
本节我们使用 89 个测试位置的 SOC 数据来验证来自 386 训练数据的普通 kriging 预测。由于我们不会使用测试数据集的 89 点来拟合模型或预测,因此这些都是对模型的独立测试。我们可以将预测值与实际值进行比较。
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
| library(plyr)
library(dplyr)
library(gstat)
library(raster)
library(ggplot2)
library(car)
library(classInt)
library(RStoolbox)
library(spatstat)
library(dismo)
library(fields)
library(gridExtra)
# Define data folder
dataFolder<-"D:\\Env\\DATA_08\\"
train<-read.csv(paste0(dataFolder,"train_data.csv"), header= TRUE)
test<-read.csv(paste0(dataFolder,"test_data.csv"), header= TRUE)
|
数据变换-幂变换
1
2
3
4
5
6
7
8
9
10
11
12
| powerTransform(train$SOC)
# Estimated transformation parameter
# train$SOC
# 0.2523339
powerTransform(test$SOC)
# Estimated transformation parameter
# test$SOC
# 0.3379903
train$SOC.bc<-bcPower(train$SOC, 0.2523339)
test$SOC.bc<-bcPower(test$SOC, 0.3379903)
|
1
2
| coordinates(train) = ~x+y
coordinates(test) = ~x+y
|
变异函数建模
1
2
3
4
5
6
7
8
9
10
| # Variogram
v<-variogram(SOC.bc~ 1, data = train, cloud=F)
# Intial parameter set by eye esitmation
m<-vgm(1.5,"Exp",40000,0.5)
# least square fit
m.f<-fit.variogram(v, m)
m.f
# model psill range
# 1 Nug 0.5165678 0.00
# 2 Exp 1.0816886 82374.23
|
测试点的预测
我们将使用 k-fold 交叉验证对模型进行评估。我们将使用 krige.cv() 函数。
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
| val<-krige(SOC.bc ~ 1,
train,
test,
model = m.f)
# [using ordinary kriging]
summary(val)
# Object of class SpatialPointsDataFrame
# Coordinates:
# min max
# x -1185457 95251.19
# y 1102846 2514145.42
# Is projected: NA
# proj4string : [NA]
# Number of points: 101
# Data attributes:
# var1.pred var1.var
# Min. :0.1426 Min. :0.7888
# 1st Qu.:1.3910 1st Qu.:0.9471
# Median :1.8303 Median :1.0099
# Mean :1.9214 Mean :1.0212
# 3rd Qu.:2.4667 3rd Qu.:1.0756
# Max. :3.8089 Max. :1.3132
|
计算残差
1
2
3
| test$SOC.pred<-val$var1.pred
test$SOC.var<-val$var1.var
test$residual<-(test$SOC.bc-test$SOC.pred)
|
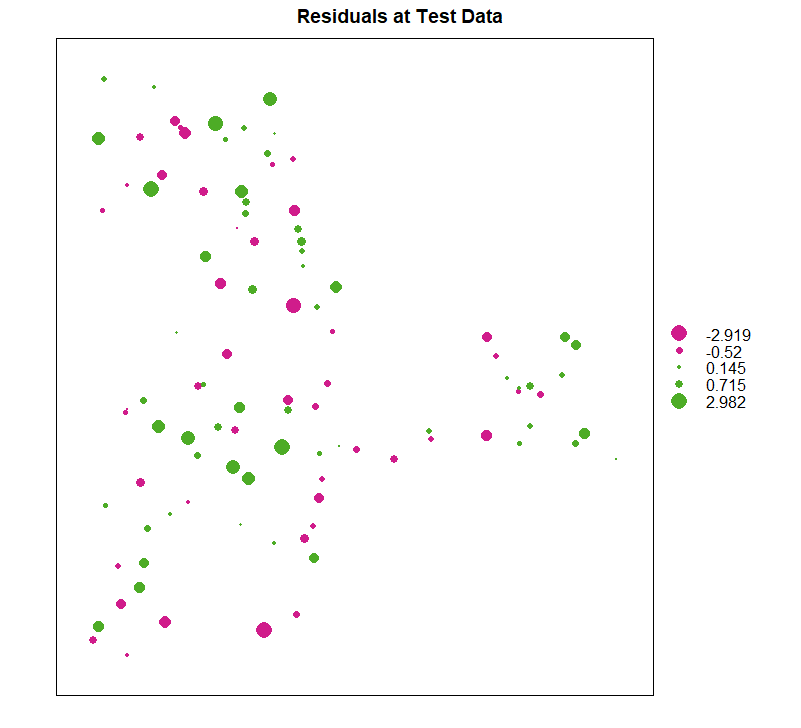
残差图
1
| bubble(test, zcol = "residual", maxsize = 2.0, main = "Residuals at Test Data")
|
误差
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
| # Mean Error (ME)
ME<-round(mean(test$residual),3)
# Mean Absolute Error
MAE<-round(mean(abs(test$residual)),3)
# Root Mean Squre Error (RMSE)
RMSE<-round(sqrt(mean(test$residual^2)),3)
# Mean Squared Deviation Ratio (MSDR)
MSDR<-mean(test$residual^2/test$SOC.var)
ME
# [1] 0.152
MAE
# [1] 0.873
RMSE
# [1] 1.126
MSDR
# [1] 1.267439
|
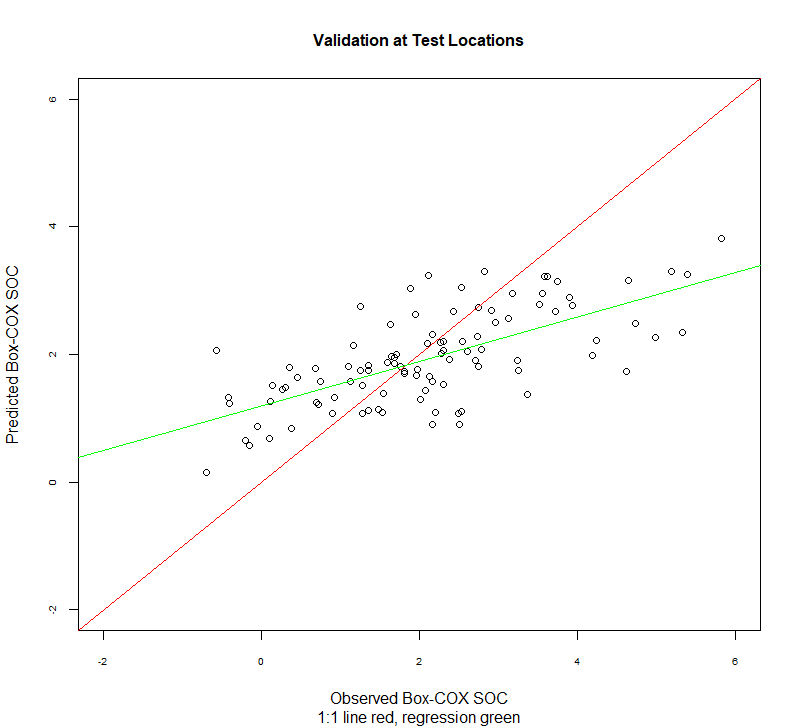
实际值 vs 预测值: 线性回归
比较实际值与预测值的另一种方法是在它们之间进行线性回归。理想情况下,这将是一条1:1 的线: 截距为0 (无偏差),斜率设置为1 (增益相等)。
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
| lm.val <- lm(test$SOC.pred ~ test$SOC.bc)
summary(lm.val)
# Call:
# lm(formula = test$SOC.pred ~ test$SOC.bc)
# Residuals:
# Min 1Q Median 3Q Max
# -1.16895 -0.42721 0.07515 0.28345 1.30121
# Coefficients:
# Estimate Std. Error t value Pr(>|t|)
# (Intercept) 1.19921 0.09196 13.041 < 2e-16 ***
# test$SOC.bc 0.34824 0.03592 9.695 5.07e-16 ***
# ---
# Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
# Residual standard error: 0.5419 on 99 degrees of freedom
# Multiple R-squared: 0.487, Adjusted R-squared: 0.4818
# F-statistic: 93.99 on 1 and 99 DF, p-value: 5.069e-16
|
1:1 图
1
2
3
4
5
6
7
8
9
10
11
| plot(test$SOC.bc, test$SOC.pred,main=list("Validation at Test Locations",cex=1),
sub = "1:1 line red, regression green",
xlab = "Observed Box-COX SOC",
ylab = "Predicted Box-COX SOC",
cex.axis=.6,
xlim = c(-2,6),
ylim =c(-2,6),
pch = 21,
cex=1)
abline(0, 1, col="red")
abline(lm.val, col = "green")
|
空间不确定性的条件模拟
该 kriging 预测图平滑了所研究属性的空间分布的局部细节,小值被高估,而大值被低估,尤其是在低采样密度的区域 (Isaaks和Srivastava 1989)。与kriging不同,条件顺序高斯模拟 (CSGS) 技术可以更好地反映现实,并消除不切实际的平滑,除了尊重原始数据值外,还着重于全局统计数据或半变异函数模型的再现 (Goovaerts 1997)。使用 csg 生成的一组替代实现提供了有关空间预测的一定程度的不确定性,该不确定性通常用于绘制所研究变量的可靠概率图 (Goovaerts 1997)。因此,csg越来越优选用于表征决策和风险分析的不确定性的 kriging,而不是像使用 kriging (Deutsch和Journel 1998) 那样产生未采样位置的最佳无偏预测。
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
| library(plyr)
library(dplyr)
library(gstat)
library(raster)
library(rasterVis)
library(ggplot2)
library(car)
library(sp)
library(classInt)
library(RStoolbox)
library(gridExtra)
dataFolder<-"D:\\Env\\DATA_08\\"
state<-shapefile(paste0(dataFolder,"GP_STATE.shp"))
train<-read.csv(paste0(dataFolder,"train_data.csv"), header= TRUE)
grid<-read.csv(paste0(dataFolder, "GP_prediction_grid_data.csv"), header= TRUE)
grid.xy<-grid[,1:3]
|
数据变换-幂变换
1
2
3
4
| powerTransform(train$SOC)
train$SOC.bc<-bcPower(train$SOC, 0.2523339)
coordinates(train) = ~x+y
coordinates(grid) = ~x+y
|
变异函数建模
1
2
3
4
5
6
7
8
9
10
| # Variogram
v<-variogram(SOC.bc~ 1, data = train, cloud=F)
# Intial parameter set by eye esitmation
m<-vgm(1.5,"Exp",40000,0.5)
# least square fit
m.f<-fit.variogram(v, m)
m.f
# model psill range
# 1 Nug 0.5165678 0.00
# 2 Exp 1.0816886 82374.23
|
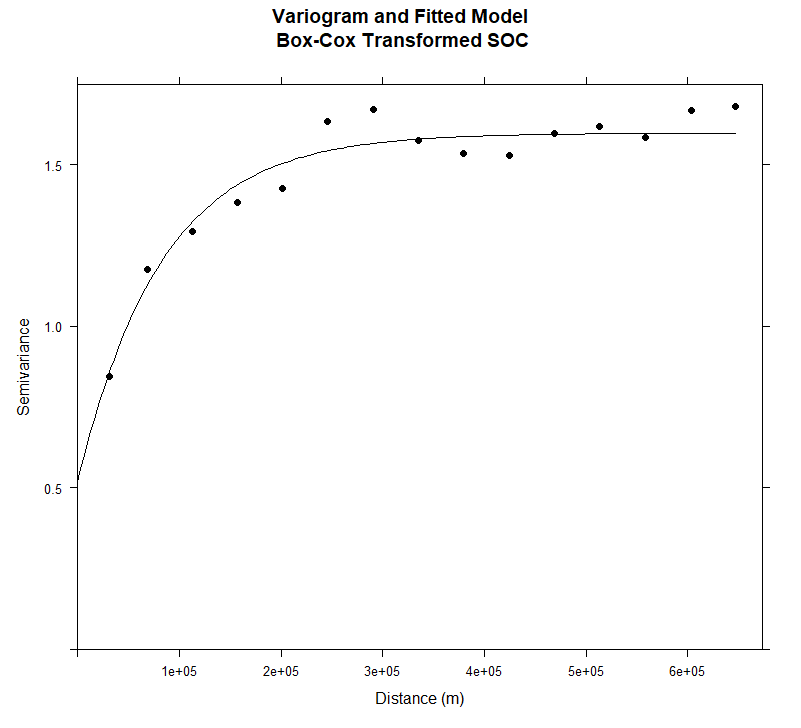
绘制变量图和拟合模型
1
2
3
4
5
6
7
8
9
10
11
| #### Plot varigram and fitted model:
plot(v, pl=F,
model=m.f,
col="black",
cex=0.9,
lwd=0.5,
lty=1,
pch=19,
main="Variogram and Fitted Model\n Box-Cox Transformed SOC",
xlab="Distance (m)",
ylab="Semivariance")
|
普通克里金
1
2
3
4
5
6
7
8
9
10
| ok.soc<-krige(SOC.bc~1,
train,
grid,
model=m.f,
nmax=50
)
# back transformation
k1<-1/0.2523339
grid.xy$OK <-((ok.soc$var1.pred *0.2523339+1)^k1)
|
条件高斯模拟
krige() 方法也可以做条件模拟。它需要一个可选的参数: nsim ,条件模拟的数量
1
2
3
4
5
6
7
8
9
10
| set.seed(130)
nsim=100
sim.soc<-krige(SOC.bc~1,
train,
grid,
model=m.f,
nmax=50,
nsim = nsim)
# drawing 100 GLS realisations of beta...
# [using conditional Gaussian simulation]
|
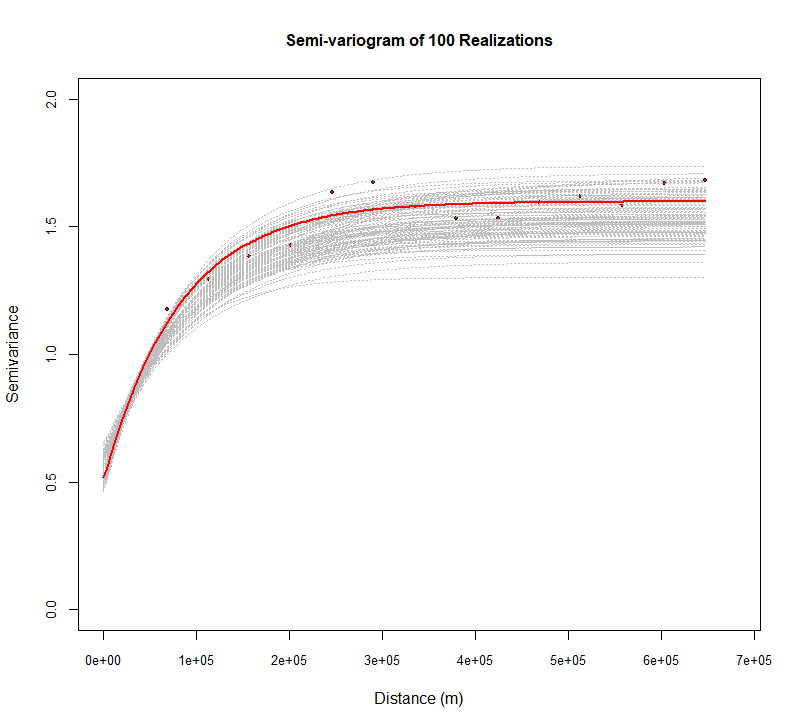
所有实现的半真实图
模拟应该尊重空间结构-与变异函数模型相同的结构。现在将产生 100 实现的变量,并绘制box-cox变换 SOC 的变量。
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
| plot(v$gamma ~ v$dist, xlim = c(0, max(v$dist) * 1.05),
ylim=c(0,2),
pch = 19, col = "black",cex=.5,
cex.axis = 0.8, cex.lab=1, xlab = list("Distance (m)",cex=1), ylab =list("Semivariance",cex=1),
main=list("Semi-variogram of 100 Realizations",cex=1))
for(i in 1:100) {
sg.v=paste("sim",i,sep="")
fg.v = as.formula(paste(sg.v, "~1"))
vg.v = variogram(fg.v, sim.soc)
lines(variogramLine(fit.variogram(vg.v, m.f),
maxdist = max(v$dist)),lty=3, col = "grey")
}
points(v$gamma ~ v$dist,pch = 19, col = "red",cex=2)
lines(variogramLine(fit.variogram(v, m.f),
maxdist = max(v$dist)), col = "red", lwd=2.0)
|
逆变换
1
2
| for(i in 1:length(sim.soc@data)){sim.soc@data[[i]] <- (((sim.soc[[i]]*0.2523339+1)^k1))}
sim.data<-as.data.frame(sim.soc)
|
空间不确定性的可视化
该组实现提供了有关砷浓度空间分布的不确定性的度量。实现之间的差异描述了不确定性。这种不确定性可以通过所有实现的动画显示来可视化,这允许区分在所有实现 (低不确定性) 上保持稳定的区域与在实现之间发生大波动的区域 (高不确定性)。
数据准备
1
2
3
4
5
6
7
8
| soc=sim.data[,3:102]
soc.stack=stack(soc)
# round(quantile(soc.stack$values, probs=seq(0,1, by=.1)),1)
at.soc=classIntervals(soc.stack$values, n = 10, style = "quantile")$brks
rgb.palette <- colorRampPalette(c("red" ,"yellow","green","blue"),space = "rgb")
bound <- list("sp.lines", as(state, "SpatialLines"), col="grey40", lwd=.8,lty=1)
coordinates(sim.data) <- ~x+y
gridded(sim.data) <- TRUE
|
要在 R 中创建动画地图,您必须在 R 中安装 animation 包。这个包依赖于 ImageMagick 软件。
1
2
3
4
| Sys.setenv(PATH = paste("C:\\Program Files\\ImageMagick-7.1.0-Q16-HDRI", Sys.getenv("PATH"), sep = ";"))
# ani.options(convert="C:\\Program Files\\ImageMagick-7.0.6-0-Q16\\covert.exe")
magickPath<-shortPathName("C:\\Program Files\\ImageMagick-7.1.0-Q16-HDRI\\magick.exe")
ani.options(convert=magickPath)
|
动画地图
1
2
3
4
5
6
7
8
9
| saveGIF(
for (i in 1:100){
print(spplot(sim.data[,i], main = list(label=paste("Realization",i),cex=1.5),
sp.layout=list(bound),
par.settings=list(axis.line=list(col="grey25",lwd=0.5)),at=at.soc,
colorkey=list(space="right",width=1.4,at=1:11,labels=list(cex=1.2,at=1:11,
labels=c("", "< 1.2", "", " 2.9", "", " 4.7", "", " 7.2", "", "> 12.5", ""))),
col.regions=rgb.palette(20)))},
height = 600, width = 600, interval = .5, outdir = getwd())
|
局部不确定性的度量
现在,我们将通过比较 E-tpye (平均值),预期 10% 概率的条件 qauntiles图,50% (中位数) 和 90% 来测量局部不确定性。
1
2
3
4
5
6
7
8
9
| # E-type estimate
df<-as.data.frame(sim.data)
df<-df[,1:100]
grid.xy$mean<-as.data.frame(rowMeans(df[sapply(df, is.numeric)]))
# names(grid.xy)[5]<-"Etype"
## Conditional Quantiles
grid.xy$q10<-apply(as.data.frame(sim.data)[3:102],1,stats::quantile,probs = 0.1,na.rm=TRUE)
grid.xy$Median<-apply(as.data.frame(sim.data)[3:102],1,stats::quantile,probs = 0.5,na.rm=TRUE)
grid.xy$q90<-apply(as.data.frame(sim.data)[3:102],1,stats::quantile,probs = 0.9,na.rm=TRUE)
|
1
2
3
4
5
6
| coordinates(grid.xy) <- ~x+y
gridded(grid.xy) <- TRUE
col <- colorRampPalette(c("red" ,"yellow","green","blue","sky blue"),space = "rgb")
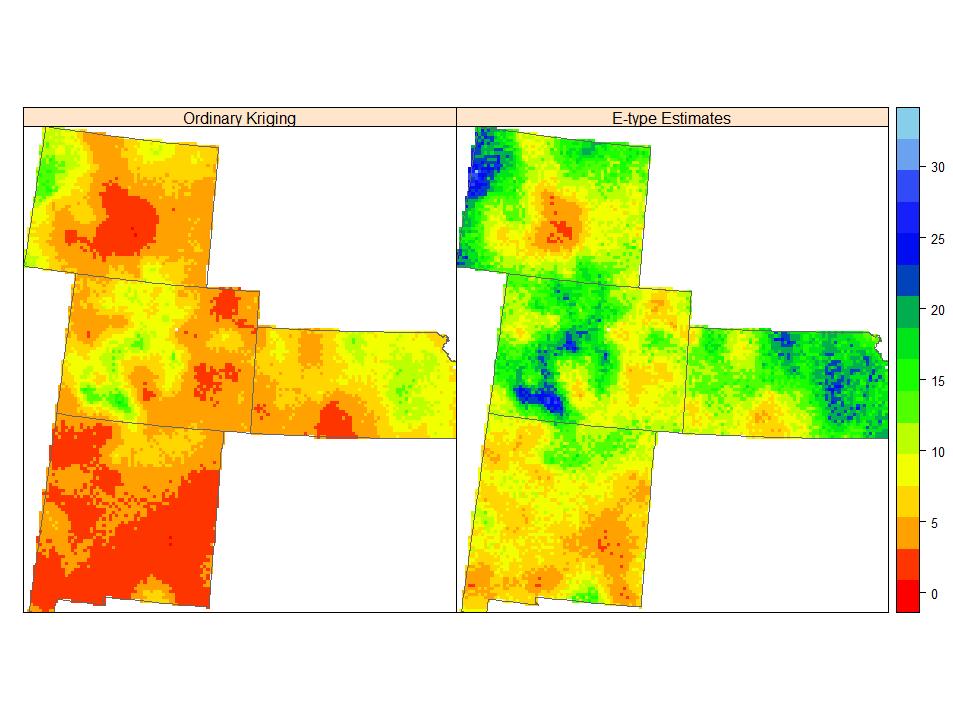
spplot(grid.xy, c(4:5), sp.layout=list(bound), col.regions=col(20),
names.attr = c("Ordinary Kriging", "E-type Estimates"))
|
100 实现的普通预测和 E 型估计并不相同,尽管两者都是最小二乘标准的最佳估计。
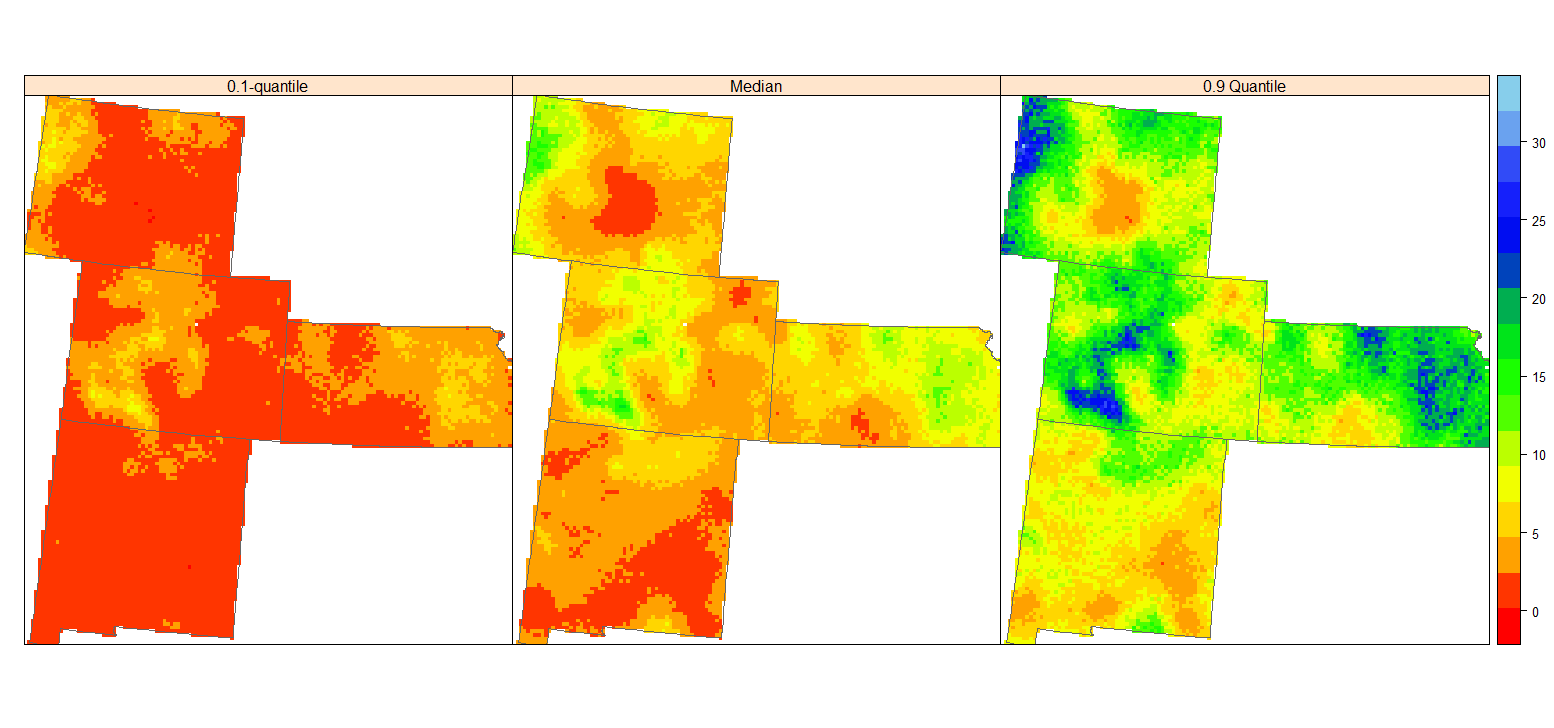
条件分位数图
1
2
| spplot(grid.xy, c("q10","Median","q90"), sp.layout=list(bound), col.regions=col(20),
names.attr = c("0.1-quantile", "Median","0.9 Quantile"), layout=c(3,1))
|
0.1 分位数图的高土壤碳 (黄色部分) 表示未知 SOC 浓度肯定较大的区域,而 0.9 图的低值部分 (深黄色) SOC 浓度肯定较小。